Detection of Major Structural Chromosomal Abnormalities in Human Karyotypes Using Convolutional Neural Networks
Ramin Yousefpour Shahrivar1, Fatemeh Karami2
Abstract
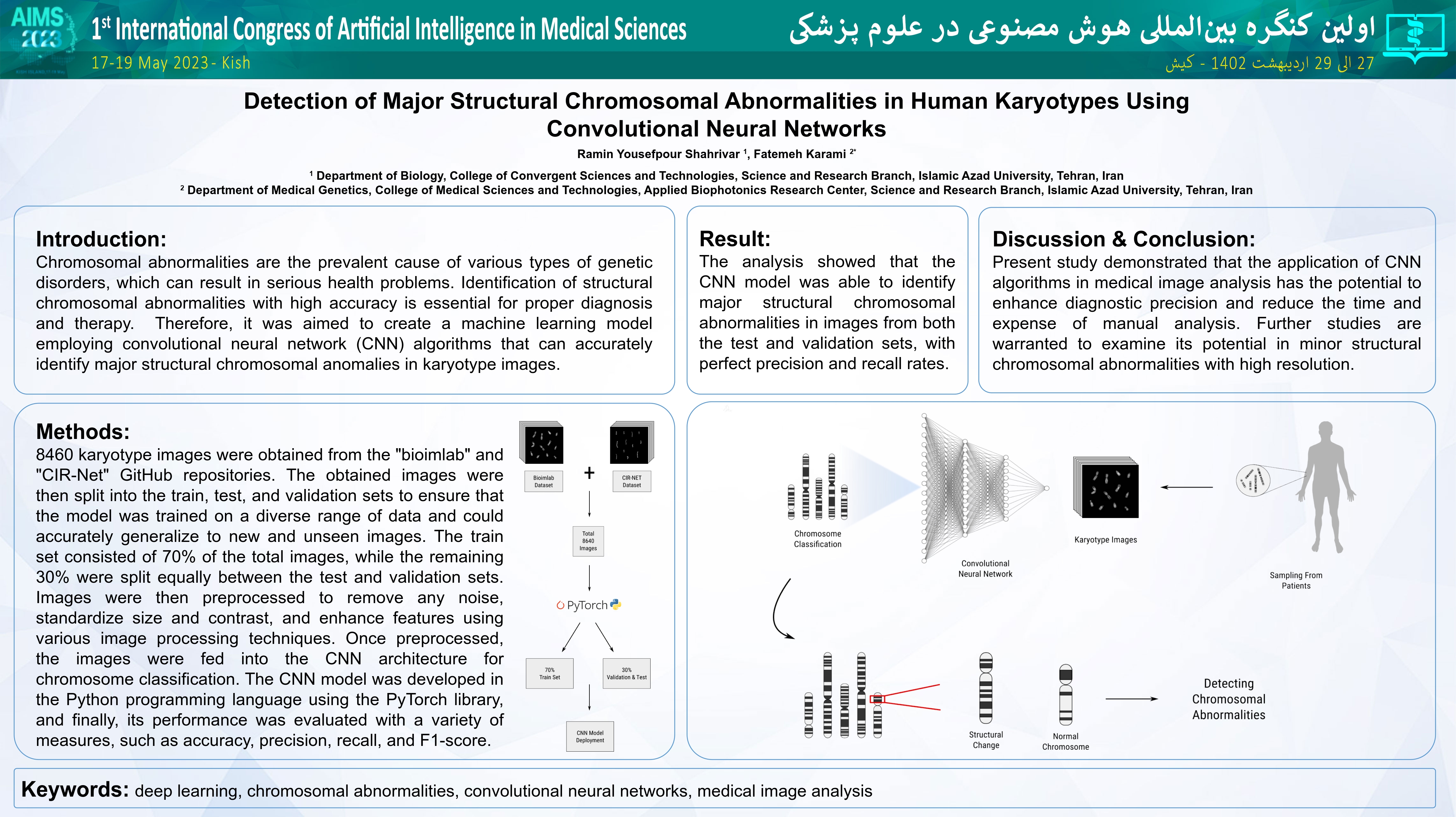
Background and aims: Chromosomal abnormalities are the prevalent cause of various types of genetic disorders, which can result in serious health problems. Identification of structural chromosomal abnormalities with high accuracy is essential for proper diagnosis and therapy. Therefore, it was aimed to create a machine learning model employing convolutional neural network (CNN) algorithms that can accurately identify major structural chromosomal anomalies in karyotype images.
Method: 8460 karyotype images were obtained from the “bioimlab” and “CIR-Net” GitHub repositories. The obtained images were then split into the train, test, and validation sets to ensure that the model was trained on a diverse range of data and could accurately generalize to new and unseen images. The train set consisted of 70% of the total images, while the remaining 30% were split equally between the test and validation sets. Images were then preprocessed to remove any noise, standardize size and contrast, and enhance features using various image processing techniques. Once preprocessed, the images were fed into the CNN architecture for chromosome classification. The CNN model was developed in the Python programming language using the PyTorch library, and finally, its performance was evaluated with a variety of measures, such as accuracy, precision, recall, and F1-score.
Results: The analysis showed that the CNN model was able to identify major structural chromosomal abnormalities in images from both the test and validation sets, with great precision and recall rates.
Conclusion: Present study demonstrated that the application of CNN algorithms in medical image analysis has the potential to enhance diagnostic precision and reduce the time and expense of manual analysis. Further studies are warranted to examine its potential in minor structural chromosomal abnormalities with high resolution. Keywords: deep learning, chromosomal abnormalities, convolutional neural networks, medical image analysis